O dimer
Overview > O atom > O atom spinpolarized > O atom spinpolarized low symmetry > O dimer > CO > CO vibration > CO partial DOS > H2O >
H2O vibration > H2O molecular dynamics > Further things to try > List of tutorials
Task
Relaxation of the bond length of an oxygen dimer.
Input
POSCAR
O dimer in a box 1.0 ! universal scaling parameters 8.0 0.0 0.0 ! lattice vector a(1) 0.0 8.0 0.0 ! lattice vector a(2) 0.0 0.0 8.0 ! lattice vector a(3) 2 ! number of atoms cart ! positions in cartesian coordinates 0 0 0 ! first atom 0 0 1.22 ! second atom
INCAR
SYSTEM = O2 dimer in a box ISMEAR = 0 ! Gaussian smearing ISPIN = 2 ! spin polarized calculation NSW = 5 ! 5 ionic steps IBRION = 2 ! use the conjugate gradient algorithm
KPOINTS
Gamma-point only 0 Monkhorst Pack 1 1 1 0 0 0
Calculation
- We have selected in the INCAR file that geometry relaxation should be performed. In this case 5 ionic steps (NSW=5) should be done at most. For the relaxation a conjugate gradient (CG) algorithm is used (IBRION=2).
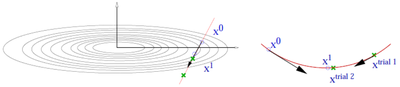
- The CG algorithm requires line minimizations along the search direction. This is done using a variant of Brent's algorithm. (Picture missing)
- Trial step along search direction (gradient scaled by POTIM)
- Quadratic or cubic interpolation using energies and forces at and allows to determine the approximate minimum
- Continue minimization, if app. minimum is not accurate enough
stdout
DAV: 1 0.517118590134E+02 0.51712E+02 -0.31393E+03 80 0.366E+02 ... ... ... ... ... ... DAV: 14 -0.985349953776E+01 -0.15177E-03 -0.57546E-06 64 0.125E-02 0.371E-03 DAV: 15 -0.985357023804E+01 -0.70700E-04 -0.22439E-06 64 0.741E-03 1 F= -.98535702E+01 E0= -.98535702E+01 d E =-.985357E+01 mag= 2.0000 curvature: 0.00 expect dE= 0.000E+00 dE for cont linesearch 0.000E+00 trial: gam= 0.00000 g(F)= 0.113E+00 g(S)= 0.000E+00 ort = 0.000E+00 (trialstep = 0.100E+01) search vector abs. value= 0.113E+00 bond charge predicted ... ... ... ... ... ... 2 F= -.96234585E+01 E0= -.96234585E+01 d E =0.230112E+00 mag= 2.0000 trial-energy change: 0.230112 1 .order 0.190722 -0.113406 0.494850 step: 0.1397(harm= 0.1864) dis= 0.00731 next Energy= -9.861386 (dE=-0.782E-02) bond charge predicted ... ... ... ... ... ... 3 F= -.98607735E+01 E0= -.98607735E+01 d E =-.720327E-02 mag= 2.0000 curvature: -0.09 expect dE=-0.900E-05 dE for cont linesearch -0.900E-05 trial: gam= 0.00000 g(F)= 0.969E-04 g(S)= 0.000E+00 ort =-0.331E-02 (trialstep = 0.828E+00) search vector abs. value= 0.969E-04 reached required accuracy - stopping structural energy minimisation
Explanation of the output:
- The quantitiy trial-energy change is the change of the energy in the trial step.
- The first value after 1. order is the expected energy change calculated from the forces change of positions - central difference.
- The second and third value correspond to change of positions and change of position.
- The value step is the estimated size of the step leading to a line minimization along the current search direction. harm is the optimal step using a second order (or harmonic) interpolation.
- The trial step sizde can be controlled by the paramter POTIM.
- The final positions after the optimization are stored in the CONTCAR file. One can copy CONTCAR to POSCAR and continue the relaxation.
Download
To the list of examples or to the main page