Liquid Si - MLFF
Task
Generating a machine learning force field for liquid Si. For this tutorial, we expect that the user is already familiar with running conventional ab initio molecular dynamic calculations.
Input
POSCAR
In this example we start from a 64 atom super cell of diamond-fcc Si (the same as in Liquid Si - Standard MD):
Si cubic diamond 2x2x2 super cell of conventional cell
5.43090000000000
2.00000000 0.00000000 0.00000000
0.00000000 2.00000000 0.00000000
0.00000000 0.00000000 2.00000000
Si
64
Direct
0.00000000 0.00000000 0.00000000
0.50000000 0.00000000 0.00000000
0.00000000 0.50000000 0.00000000
0.50000000 0.50000000 0.00000000
0.00000000 0.00000000 0.50000000
0.50000000 0.00000000 0.50000000
0.00000000 0.50000000 0.50000000
0.50000000 0.50000000 0.50000000
0.37500000 0.12500000 0.37500000
0.87500000 0.12500000 0.37500000
0.37500000 0.62500000 0.37500000
0.87500000 0.62500000 0.37500000
0.37500000 0.12500000 0.87500000
0.87500000 0.12500000 0.87500000
0.37500000 0.62500000 0.87500000
0.87500000 0.62500000 0.87500000
0.00000000 0.25000000 0.25000000
0.50000000 0.25000000 0.25000000
0.00000000 0.75000000 0.25000000
0.50000000 0.75000000 0.25000000
0.00000000 0.25000000 0.75000000
0.50000000 0.25000000 0.75000000
0.00000000 0.75000000 0.75000000
0.50000000 0.75000000 0.75000000
0.37500000 0.37500000 0.12500000
0.87500000 0.37500000 0.12500000
0.37500000 0.87500000 0.12500000
0.87500000 0.87500000 0.12500000
0.37500000 0.37500000 0.62500000
0.87500000 0.37500000 0.62500000
0.37500000 0.87500000 0.62500000
0.87500000 0.87500000 0.62500000
0.25000000 0.00000000 0.25000000
0.75000000 0.00000000 0.25000000
0.25000000 0.50000000 0.25000000
0.75000000 0.50000000 0.25000000
0.25000000 0.00000000 0.75000000
0.75000000 0.00000000 0.75000000
0.25000000 0.50000000 0.75000000
0.75000000 0.50000000 0.75000000
0.12500000 0.12500000 0.12500000
0.62500000 0.12500000 0.12500000
0.12500000 0.62500000 0.12500000
0.62500000 0.62500000 0.12500000
0.12500000 0.12500000 0.62500000
0.62500000 0.12500000 0.62500000
0.12500000 0.62500000 0.62500000
0.62500000 0.62500000 0.62500000
0.25000000 0.25000000 0.00000000
0.75000000 0.25000000 0.00000000
0.25000000 0.75000000 0.00000000
0.75000000 0.75000000 0.00000000
0.25000000 0.25000000 0.50000000
0.75000000 0.25000000 0.50000000
0.25000000 0.75000000 0.50000000
0.75000000 0.75000000 0.50000000
0.12500000 0.37500000 0.37500000
0.62500000 0.37500000 0.37500000
0.12500000 0.87500000 0.37500000
0.62500000 0.87500000 0.37500000
0.12500000 0.37500000 0.87500000
0.62500000 0.37500000 0.87500000
0.12500000 0.87500000 0.87500000
0.62500000 0.87500000 0.87500000
KPOINTS
We will start with a single k point in this example:
K-Points 0 Gamma 1 1 1 0 0 0
INCAR
#Basic parameters ISMEAR = 0 SIGMA = 0.1 LREAL = Auto ISYM = -1 NELM = 100 EDIFF = 1E-4 LWAVE = .FALSE. LCHARG = .FALSE. #Parallelization of ab initio calculations NCORE = 2 #MD IBRION = 0 MDALGO = 2 ISIF = 2 SMASS = 1.0 TEBEG = 2000 NSW = 10000 POTIM = 3.0 #Machine learning paramters ML_FF_LMLFF = .TRUE. ML_FF_ISTART = 0 ML_FF_EATOM = -.70128086E+00
- ML_FF_LMLFF = .TRUE.
- switches on the machine learning of the force field
- ML_FF_ISTART = 0
- corresponds to learning from scratch
- ML_FF_EATOM = -.70128086E+00
- sets the atomic reference energy for each species. We show to obtain this energy below.
- IWAVPR = 1
- extrapolate the total charge density from the fixed atomic ones. This is more advisable than the default wavefunction-based extrapolation from previous steps, because during the on-the-fly learning, several thousands of molecular dynamics steps can lie between two ab initio calculations.
Calculation
Reference energy
To obtain the force field for liquid Si, we require the atomic energy of a single Si atom. We create a new working directory for this calculation
mkdir Si_ATOM cd Si_ATOM ln -s ../POTCAR
In this directory, we create the following INCAR file
#Basic parameters ISMEAR = 0 SIGMA = 0.1 LREAL = Auto ISYM = 0 NELM = 100 EDIFF = 1E-4 LWAVE = .FALSE. LCHARG = .FALSE. ISPIN = 2
The POSCAR file contains a single atom in a large enough box (the box should be orthorhombic to have enough degrees of freedom for electronic relaxation)
Si atom
1.00090000000000
12.00000000 0.00000000 0.00000000
0.00000000 12.01000000 0.00000000
0.00000000 0.00000000 12.02000000
Si
1
Direct
0.00000000 0.00000000 0.00000000
Due to the large unit cell, we can use a Gamma point calculation set up in the KPOINTS file
Gamma point only 0 0 0 Gamma 1 1 1 0 0 0
After running the calculation, we obtain the total energy from the OUTCAR or OSZICAR file. We use this energy for the ML_FF_EATOM tag. If multiple atom types are present in the structure, this step has to be repeated for each atom type separately and the reference energies are provided as a list after the ML_FF_EATOM tag. The ordering of the energies corresponds to the ordering of the elements in the POTCAR file.
Creating the liquid structure
Because we don't have a structure of liquid silicon readily available, we first create that structure by starting from a super cell of crystalline silicon with 64 atoms. The temperature is set to 2000 K so that the crystal melts rapidly in the MD run. To improve the simulation speed drastically, we utilize the on-the-fly machine learning. Most of the ab initio steps will be replaced by very fast force-field ones. Within 10000 steps equivalent to 30 ps, we have obtained a good starting position for the subsequent simulations in the CONTCAR file. You can copy the input files or download them.
After running the calculation, we obtained a force field, but its initial trajectory might be tainted but the unreasonable starting position. Nevertheless, it is instructive to inspect the output to understand how to assess the accuracy of a force field, before refining it in subsequent calculations. The main output files for the machine learning are
- ML_ABNCAR
- contains the ab initio structure datasets used for the learning. It will be needed for continuation runs as ML_ABCAR.
- ML_FFNCAR
- contains the regression results (weights, parameters, etc.). It will be needed for continuation runs as ML_FFCAR.
- ML_LOGFILE
- logging the proceedings of the machine learning. With ML_FF_NWRITE = 2 set, this file lists the error between force-field and ab initio calculation for energy, forces and the stress. The entry for the last step should be similar to the following (the inherent randomness of a molecular dynamics simulation will lead to slightly different results):
====================================================================================================
Information on error estimations
----------------------------------------------------------------------------------------------------
Error in energy (eV atom^-1): 0.016994
Error in force (eV Angst^-1): 0.247664
Error in stress (kB): 5.368218
Bayesian error (eV Angst-1): 0.064904 0.127195
Spilling factor (-): 0.000690 0.020000
====================================================================================================
You can see that additional information about the Bayesian error and the spilling factor is reported. The first entry of the Bayesian error is the estimated error from our model. If the estimated error exceeds the threshold listed next to it a new structure dataset is generated. The threshold is automatically determined during the calculations. The first and second entry for the spilling factor are the calculated spilling factor and the threshold, respectively. The threshold for the spilling factor is usually kept constant during the calculations.
Structral properties of the force field
To examine the accuracy of structural properties, we compare the deviations between a 3 ps molecular dynamics run using the force field and a full ab initio calculation. For a meaningful comparison, it is best to start from the same initial structure. We will use the liquid structure, we obtained in the previous step and back it up
cp CONTCAR POSCAR.T2000_relaxed
Now, we proceed with the force field calculation and set up the required files
cp POSCAR.T2000_relaxed POSCAR cp ML_ABNCAR ML_ABCAR cp ML_FFNCAR ML_FFCAR
To run a shorter simulation using only the force field, we change the following INCAR tags to
ML_FF_ISTART = 2 NSW = 1000
After the calculation finished, we backup the history of the atomic positions
cp XDATCAR XDATCAR.MLFF_3ps
We repeat the same process for the ab initio simulation recovering the same initial structure
cp POSCAR.T2000_relaxed POSCAR
switching the machine learning off in the INCAR file by setting
ML_FF_LMLFF = .FALSE.
We store the trajectory once the calculation completed
cp XDATCAR XDATCAR.AI_3ps
To analyze the pair correlation function, we use the PERL script pair_correlation_function.pl
and process the previously saved XDATCAR files
perl pair_correlation_function.pl XDATCAR.MLFF_3ps > pair_MLFF_3ps.dat perl pair_correlation_function.pl XDATCAR.AI_3ps > pair_AI_3ps.dat
We can compare the pair correlation functions, e.g. with gnuplot using the following command
gnuplot -e "set terminal jpeg; set xlabel 'r(Ang)'; set ylabel 'PCF'; set style data lines; plot 'pair_MLFF_3ps.dat', 'pair_AI_3ps.dat' " > PC_MLFF_vs_AI_3ps.jpg
The pair correlation functions obtained that way should look similar to this figure
We see that pair correlation is quite well reproduced although the error on the force is a little bit large. This demonstrates a drastic improvement of the time consuming melting a crystal by machine learning and maybe useful even in situations where full ab initio accuracy for the liquid MD is required, because the initial equilibration phase is accelerated.
Obtaining a more accurate force field
Including the melting phase in the force field may impact the accuracy of the force field. To improve it is usually advisable to learn on the pure structures, which in our case this means to use the CONTCAR file obtained after the melting. Furthermore, the force field was learned using only a single k point so that we can improve the accuracy of the reference data by including more k points. In most calculations, it is important to conduct accurate ab initio calculations since bad reference data can limit the accuracy or even inhibit the learning of a force field.
We restart from the liquid structure obtained before
cp POSCAR.T2000_relaxed POSCAR
and change the following INCAR tags
ALGO = Normal ML_FF_LMLFF = .TRUE. ML_FF_ISTART = 0 NSW = 10000
If you run have resources to run in parallel, you can reduce the computation time further by adding k point parallelization with the KPAR tag. We use a denser k-point mesh in the KPOINTS file
2x2x2 Gamma-centered mesh 0 0 0 Gamma 2 2 2 0 0 0
We will learn a new force field on a run of 30 ps. Keep in mind to run the calculation using the standard version of VASP (usually vasp_std). After running the calculation, we examine the error in the ML_LOGFILE, where the obtained values should be close to
====================================================================================================
Information on error estimations
----------------------------------------------------------------------------------------------------
Error in energy (eV atom^-1): 0.006079
Error in force (eV Angst^-1): 0.165678
Error in stress (kB): 2.553148
Bayesian error (eV Angst-1): 0.044695 0.101357
Spilling factor (-): 0.002285 0.020000
====================================================================================================
We immediately see that the errors are significantly lower than in the previous calculation with only one k point. This is due to the less noisy ab initio data which is easier to learn.
To understand how the force field is learned, we inspect the ML_ABNCAR file containing the training data. In the beginning of this file, you will find information about the number of reference structures
**************************************************
The number of configurations
--------------------------------------------------
55
and the size of the basis set
**************************************************
The numbers of basis sets per atom type
--------------------------------------------------
396
We will monitor how much training data is added to a structure after each learning cycle and what impact this has on the accuracy of the force field. First we will perform a continuation run keeping the Bayesian threshold fixed
cp ML_ABNCAR ML_ABCAR cp CONTCAR POSCAR
and set the following INCAR tags
ML_FF_ISTART = 1 ML_FF_CTIFOR = x
where x is the threshold of the Bayesian error obtained from the ML_LOGFILE (highlighted bold above; please use the value obtained in your calculation). By setting a good estimate for the threshold several ab initio steps are skipped in the first few steps, which would be required to determine the threshold automatically. Mind: When continuation runs are performed for different crystal structures, the last previous threshold for the Bayesian error might be too large leading to premature skipping of ab initio steps. This is particular relevant when studying the liquid phase, first, and applying its threshold to the solid phase. In that case it is safer to use the default value for ML_FF_CTIFOR = .
After running the calculation, we inspect the last instance of the errors in the ML_LOGFILE, which should look similar to
====================================================================================================
Information on error estimations
----------------------------------------------------------------------------------------------------
Error in energy (eV atom^-1): 0.006252
Error in force (eV Angst^-1): 0.168549
Error in stress (kB): 2.606332
Bayesian error (eV Angst-1): 0.037569 0.101357
Spilling factor (-): 0.001114 0.020000
====================================================================================================
We see that the accuracy has not changed significantly and the threshold for the Bayesian error is set to the one specified in the INCAR file. Looking at the actual errors we observe that the threshold was exceeded only a few times. You can find all these instances with the command
grep "Bayesian error (eV Angst-1):" ML_LOGFILE | awk '{if ($5 > $6) { print $5 }}'
Consequently only very few structures were added to the ab initio data in the ML_ABNCAR file. The entry for the reference structures and basis sets should look similar to
**************************************************
The number of configurations
--------------------------------------------------
65
...
**************************************************
The numbers of basis sets per atom type
--------------------------------------------------
407
From this observation, we conclude either that the force field is already accurate enough or that the threshold is set too high. In the latter case, the machine would not conduct ab initio calculations because the force field is judged accurate enough according to the threshold.
We compare this behavior to the case of using the default value for ML_FF_CTIFOR. We continue from the existing structure and force field
cp CONTCAR POSCAR cp ML_ABNCAR ML_ABCAR
Remove the ML_FF_CTIFOR tag from the INCAR file.
The default value for ML_FF_CTIFOR is practically zero, so we expect more structures to be added. We inspect the ML_ABNCAR to investigate how the number of structures and the basis set size changed
**************************************************
The number of configurations
--------------------------------------------------
160
...
**************************************************
The numbers of basis sets per atom type
--------------------------------------------------
726
Even though the number of structure datasets and the basis set size increased, the accuracy of the force field did not improve significantly
====================================================================================================
Information on error estimations
----------------------------------------------------------------------------------------------------
Error in energy (eV atom^-1): 0.005950
Error in force (eV Angst^-1): 0.162064
Error in stress (kB): 2.628567
Bayesian error (eV Angst-1): 0.029723 0.061348
Spilling factor (-): 0.000406 0.020000
====================================================================================================
Ideally, one should continue learning until no structures need to be added to the training data and basis set anymore. In practice, the prediction of the Bayesian error exhibits numerical inaccuracies so that an ab initio calculation is conducted even if the force field is accurate enough. Hence, in this tutorial, we stop at this point because further addition of training data does not improve the accuracy on the test structures compared to the ab initio data. For high quality production calculations suited for publications, we advise to learn until almost no data is added to the training structures and basis set. In many cases this involves a runtime of at least 100 ps for learning.
- Exercise
- Continue learning and observe the convergence of the number of reference structures. Most likely the maximum allowed basis set size ML_FF_MB_MB needs to be increased. How does the pair correlation function compare to ab initio data?
Optimizing weights of energy, forces and stress
Even after a force field has been learned, there is still the possibility to refine it by changing the weights for energy ML_FF_WTOTEN, forces ML_FF_WTIFOR and stress ML_FF_WTSIF. By default all weights are set to 1.0, so energy, force and stress are equally important. Typically the error for a component decreases with increasing weight of that component. For methods where the free energy is important (such as e.g. Interface pinning calculations, Metadynamics etc.) introducing a larger weight for the energy can reduce the error of the energy significantly while only slightly increasing the errors of the forces and the stress. Note that the weights are set relative to each other, i.e., by increasing one the other ones decrease automatically.
We will start from the force field obtained above, but you can use a more refined one if you completed the user task
cp INCAR INCAR.run cp CONTCAR POSCAR.run
The INCAR.run file will serve as a template for several short calculations with different weights for the energy. Set
ML_FF_ISTART = 1 NSW = 1
to generate a single new test structure.
We can now use a script to set ML_FF_WTOTEN to different values and examine how the average error changes
#!/bin/bash
if [ -f error_vs_eweight.dat ]; then
rm error_vs_eweight.dat
fi
touch error_vs_eweight.dat
for i in 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0
do
cat INCAR.run > INCAR
echo "ML_FF_WTOTEN = $i" >> INCAR
cp POSCAR.run POSCAR
mpirun -np $np $executable_path/vasp_std
cp ML_LOGFILE ML_LOGFILE.$i
awk -v var="$i" '/Error in energy \(eV atom\^-1\)\:/{energy=$6} /Error in force \(eV Angst\^-1\)\:/ {force=$6} /Error in stress \(kB\)\:/{stress=$5} /Energy SD. \(eV atom\^-1\)\:/{energy_var=$5} /Force SD. \(eV Angst\^-1\)\:/{force_var=$5} /Stress SD. \(kB\)\:/ {stress_var=$4} END{print var,energy/energy_var+force/force_var+stress/stress_var}' ML_LOGFILE.$i >> error_vs_eweight.dat
done
To run this script you need to
export executable_path=path to the vasp executable export np=number of processes you want to use
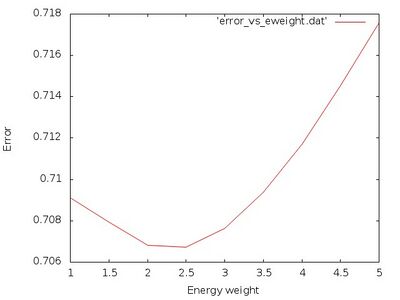
This script sets the weight of the energy ML_FF_WTOTEN to values between 1.0 and 5.0. It calculates a single structure and compares the error of energy, forces and stress between ab initio calculation and force field. These errors are normalized with respect to the standard deviations of these quantities. The sum of the normalized errors is plotted against the weight of the energy with the following command
gnuplot -e "set terminal jpeg; set xlabel 'Energy weight'; set ylabel 'Error'; set style data lines; plot 'error_vs_eweight.dat'" > error_vs_eweight.jpg
It should look similar to
From this plot we see that an ideal value is around ML_FF_WTOTEN=2.5. Next we take a look at the actual errors for that weighting parameter in the file ML_LOGFILE.2.5:
====================================================================================================
Information on error estimations
----------------------------------------------------------------------------------------------------
Error in energy (eV atom^-1): 0.005322
Error in force (eV Angst^-1): 0.169356
Error in stress (kB): 2.849817
Bayesian error (eV Angst-1): 0.025548 0.000000
Spilling factor (-): 0.000368 0.020000
====================================================================================================
We see that the error on energy decreased while it increased for forces and stress compared to ML_LOGFILE.1.0:
====================================================================================================
Information on error estimations
----------------------------------------------------------------------------------------------------
Error in energy (eV atom^-1): 0.005954
Error in force (eV Angst^-1): 0.162006
Error in stress (kB): 2.628128
Bayesian error (eV Angst-1): 0.028542 0.000000
Spilling factor (-): 0.000368 0.020000
====================================================================================================
In this example the decrease of the energy is only small but in many examples this can lead to significant increase in the precision. Compare these values to the errors in ML_LOGFILE_5.0:
====================================================================================================
Information on error estimations
----------------------------------------------------------------------------------------------------
Error in energy (eV atom^-1): 0.004692
Error in force (eV Angst^-1): 0.179448
Error in stress (kB): 3.180711
Bayesian error (eV Angst-1): 0.023225 0.000000
Spilling factor (-): 0.000368 0.020000
====================================================================================================
We see that for higher values of the weight the energy is even more accurate, but at the expense of the forces and the stress. Consequently the normalized error increases so that the trajectories during the MD simulation may suffer. We advise not to push the weight of the energy to these extreme values and instead use the value optimizing the overall error.
If needed this weighting can be done in a similar way for the forces and stress.
Production runs using the optimized parameters
In the production run, we rely on the structure dataset and the corresponding accurate force field
cp CONTCAR POSCAR cp ML_ABNCAR ML_ABCAR. cp ML_FFNCAR ML_FFCAR.
In the INCAR file, we switch of the updating of the force field and use the optimized energy
ML_FF_ISTART = 2 ML_FF_WTOTEN = 2.5
and adjust the necessary molecular dynamics parameters (like e.g. NSW etc.).
- Exercise
- Calculate the pair correlation function as above with the machine learned force field and compare to results obtained ab initio when using the same POSCAR.