Category:Biased molecular dynamics: Difference between revisions
(→Output) |
|||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
''Biased molecular dynamics''' (MD) refers to advanced [[:Category:Molecular dynamics|MD-simulation methods]] that introduce a ''bias potential''. One of the most important purposes of using bias potentials is to enhance the sampling of phase space with low probability density (e.g., transition regions of chemical reactions). Depending on the type of sampling and in combination with the corresponding statistical methods one then has access to important thermodynamic quantities like, e.g., free energies. Biased molecular dynamics comes in very different flavors such as, e.g., umbrella sampling{{cite|torrie:jcp:1977}} and umbrella integration{{cite|kaestner:jcp:2005}}, to name a few. For a comprehensive description (especially about umbrella sampling), we refer the interested user to Ref. {{cite|frenkel:ap-book:2002}} written by D. Frenkel and B. Smit. | |||
The probability density for a geometric parameter ξ of the system driven by a Hamiltonian | The probability density for a geometric parameter ξ of the system driven by a Hamiltonian | ||
| Line 135: | Line 133: | ||
== Related methods in VASP == | == Related methods in VASP == | ||
*[[ | *[[Metadynamics]]: In contrast to the methods discussed on this page metadynamics continuously updates the bias potential of the system to push it into unvisited parts of phase space. | ||
*[[:Category:Interface pinning|Interface pinning]]: This employs a bias potential to pin the state of an interface between a solid and a liquid. This method uses entirely different {{TAG|INCAR}} tags than the bias potentials presented on this page. | *[[:Category:Interface pinning|Interface pinning]]: This employs a bias potential to pin the state of an interface between a solid and a liquid. This method uses entirely different {{TAG|INCAR}} tags than the bias potentials presented on this page. | ||
| Line 142: | Line 140: | ||
<references/> | <references/> | ||
[[Category: | [[Category:Advanced molecular-dynamics sampling]][[Category:Theory]] | ||
Latest revision as of 12:37, 29 October 2024
Biased molecular dynamics' (MD) refers to advanced MD-simulation methods that introduce a bias potential. One of the most important purposes of using bias potentials is to enhance the sampling of phase space with low probability density (e.g., transition regions of chemical reactions). Depending on the type of sampling and in combination with the corresponding statistical methods one then has access to important thermodynamic quantities like, e.g., free energies. Biased molecular dynamics comes in very different flavors such as, e.g., umbrella sampling[1] and umbrella integration[2], to name a few. For a comprehensive description (especially about umbrella sampling), we refer the interested user to Ref. [3] written by D. Frenkel and B. Smit.
The probability density for a geometric parameter ξ of the system driven by a Hamiltonian
with T(p), and V(q) being kinetic, and potential energies, respectively, can be written as
The term stands for a thermal average of quantity X evaluated for the system driven by the Hamiltonian H.
If the system is modified by adding a bias potential acting on one or multiple selected internal coordinates of the system ξ=ξ(q), the Hamiltonian takes the form
and the probability density of ξ in the biased ensemble is
It can be shown that the biased and unbiased averages are related via
More generally, an observable
can be expressed in terms of thermal averages within the biased ensemble as
One of the most popular methods using bias potentials is umbrella sampling[1]. This method uses a bias potential to enhance sampling of ξ in regions with low P(ξi), e.g., transition regions of chemical reactions. The correct distributions are recovered afterward using the equation for above.
How to
- First one needs to setup a standard molecular dynamics run. The bias potentials are supported in both the NVT and NpT MD simulations, regardless of the particular thermostat and/or barostat setting.
Mind: Mind that for VASP 5.x the biased molecular dynamics runs have to be chosen by adding 10 to the chosen value of MDALGO. E.g. MDALGO=12 instead of MDALGO=2 has to be chosen for Nose-Hoover thermostat.
- Then one needs to set the geometric parameters and bias potential types. The geometric parameters ξ, also called collective variables, that are controlled via the potentials are defined in the ICONST file. The type of bias potential is also set in this file. The format of this file follows this layout:
flag item(1) ... item(N) status
Here flag specifies the type of geometric parameters (bond lengths, angles, etc.), item(1) ... item(N) the actual geometric items (atom numbers, etc.) and the status sets the type of used bias potential. Here we advise the user to also look into the description of the full capabilities of the geometric parameters.
- To avoid the update of the bias potential HILLS_BIN=NSW is set by default.
- Next one needs to set the parameters of the potential in the INCAR (harmonic and step function) file or in the PENALTYPOT file (Gaussian). The following table summarizes the available potentials with their corresponding parameters:
Potential type statusin ICONSTINCAR parameters Harmonic potential 8 SPRING_K, SPRING_R0, optional SPRING_V0 Step function 4 FBIAS_A, FBIAS_D, and FBIAS_R0 Gaussian potential 5 parameters set in PENALTYPOT file
Available potentials
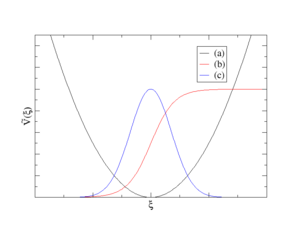
In the following details are given on how to control the available bias potentials in VASP that are plotted in Fig.1.
Harmonic potentials
- A sum of Harmonic potentials (curve (a) in Fig.1)
- where the sum runs over all () coordinates the potential acts upon. The potential is chosen in the ICONST file by setting the
statusto 8. The parameters of the potential are the force constant (SPRING_K) and the minimum or the potential SPRING_R0. These must be set in the INCAR file. Optionally, it is possible to change the value of every MD step at a constant rate defined via the INCAR tag SPRING_V0.
Mind: The number of items defined via SPRING_K, SPRING_R0, and SPRING_V0 must be equal to . Otherwise, the calculation terminates with an error message.
- This form of bias potential is employed in several simulation protocols, such as the umbrella sampling[1], umbrella integration, or steered MD, and is useful also in cases where the values need to be restrained.
Step function
- A sum of Fermi-like step functions (curve (b) in Fig.1)
- where the sum runs over all () coordinates the potential acts upon. The potential is chosen in the ICONST file by setting the
statusto 4. The parameters of the potential are the height of the step ( set by FBIAS_A), the slope around the point ( set by FBIAS_D), and the position of the step ( set by FBIAS_R0). These must be set in the INCAR file.
Mind: The number of items defined via FBIAS_A, FBIAS_D, and FBIAS_R0 must be equal to . Otherwise, the calculation terminates with an error message.
- This form of potential is suitable especially for imposing restrictions on the upper (or lower) limit of the value of .
Gaussian potential
- A sum of Gauss functions (curve (b) in Fig.1)
- where is the number of Gaussian functions and is the number of coordinates the potential acts upon. The potential is chosen in the ICONST file by setting the
statusto 5. The parameters of the potentials, , , and are defined in the PENALTYPOT file.
- This type of bias potential is primarily intended for use in metadynamics, but since Gaussians can be used as basis functions for more general shapes, they can also be used to prepare various atypically shaped bias potentials.
Output
- The values of all collective variables defined in the ICONST file for each MD step are listed in the REPORT file. Check the lines after the string Metadynamics.
Examples of usage
Let us consider the nucleophile substitution reaction of CHCl with Cl. The reactant is a weak van-der-Waals complex. The corresponding POSCAR file reads
vdW complex CH3Cl...Cl 1.00000000000000 12.0000000000000000 0.0000000000000000 0.0000000000000000 0.0000000000000000 12.0000000000000000 0.0000000000000000 0.0000000000000000 0.0000000000000000 12.0000000000000000 C H Cl 1 3 2 cart 5.91331371 7.11364924 5.78037960 5.81982231 8.15982106 5.46969017 4.92222130 6.65954232 5.88978969 6.47810398 7.03808479 6.71586385 4.32824726 8.75151396 7.80743202 6.84157897 6.18713289 4.46842049
Due to the weak interactions between CHCl and Cl, the complex can collapse at high temperatures. This can be avoided by setting an upper bound for the length of the non-bonding Cl...C interactions. This can be conveniently achieved by using a Fermi-like step-shaped bias potential. To this end, we need to define the Cl...C distance, i.e., the distance between the atoms 1 and 5, as a coordinate with status 4 in the ICONST file:
R 1 5 4
Next, we need to set the molecular dynamics parameters and specify the bias potential parameters FBIAS_A, FBIAS_D, and FBIAS_R0 in the INCAR file:
# Molecular dynamics part IBRION = 0 TEBEG = 300 TEEND = 300 MDALGO = 2 POTIM = 2.0 NSW = 10000 # Bias potential part FBIAS_A = 1 FBIAS_D = 50 FBIAS_R0 = 3.5
Since the bias potential acts only on one internal coordinate (), we need to provide only one number for each of the tags. The chosen bias potential parameters ensure that repulsive bias forces steeply increase when the C...Cl distance is increased beyond about . This causes a shortening of the distance in the next MD step. Notice that the bias force is essentially negligible for distances below . A careful adjustment of FBIAS_A and FBIAS_D is needed to ensure that (i) the bias force is large enough to effectively limit the value of , and (ii) the interval of values for which the bias forces are significant is broad enough to avoid overcoming via random fluctuations. A suitable setting can be found by noting that the maximal bias force of is exerted on the system at the point . This can be seen by inspecting the analytical expression for the potential.
Related methods in VASP
- Metadynamics: In contrast to the methods discussed on this page metadynamics continuously updates the bias potential of the system to push it into unvisited parts of phase space.
- Interface pinning: This employs a bias potential to pin the state of an interface between a solid and a liquid. This method uses entirely different INCAR tags than the bias potentials presented on this page.
References
- ↑ a b c G. M. Torrie and J. P. Valleau, J. Comp. Phys. 23, 187 (1977).
- ↑ J. Kästner, and W. Thiel, Bridging the gap between thermodynamic integration and umbrella sampling provides a novel analysis method: “Umbrella integration”, J. Chem. Phys. 123, 144104 (2005).
- ↑ D. Frenkel and B. Smit, Understanding molecular simulations: from algorithms to applications, Academic Press: San Diego, 2002.
This category currently contains no pages or media.
