CO partial DOS: Difference between revisions
Vaspmaster (talk | contribs) (Created page with '*INCAR SYSTEM = CO molecule in a box ISMEAR = 0 ! Gaussian smearing LORBIT = 11 *KPOINTS Gamma-point only 1 ! one k-point rec ! in units of the reciprocal lat…') |
|||
| (22 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Template:At_and_mol - Tutorial}} | |||
== Task == | |||
Calculation of the DOS of a CO molecule (using p4vasp) | |||
CO molecule in a box | |||
== Input == | |||
=== {{TAG|POSCAR}} === | |||
CO molecule in a box | |||
1 1 ! number of atoms for each species | 1.0 ! universal scaling parameters | ||
sel ! selective degrees of freedom are changed | 8.0 0.0 0.0 ! lattice vector a(1) | ||
cart ! positions in cartesian coordinates | 0.0 8.0 0.0 ! lattice vector a(2) | ||
0.0 0.0 8.0 ! lattice vector a(3) | |||
1 1 ! number of atoms for each species | |||
sel ! selective degrees of freedom are changed | |||
cart ! positions in cartesian coordinates | |||
0 0 0 F F T ! first atom | |||
0 0 1.143 F F T ! second atom | |||
=== {{TAG|INCAR}} === | |||
{{TAGBL|SYSTEM}} = CO molecule in a box | |||
{{TAGBL|ISMEAR}} = 0 ! Gaussian smearing | |||
{{TAGBL|LORBIT}} = 11 | |||
=== {{TAG|KPOINTS}} === | |||
Gamma-point only | |||
0 | |||
Monkhorst Pack | |||
1 1 1 | |||
0 0 0 | |||
== Calculation == | |||
*The {{TAG|PROCAR}} file gives valuable information of the character of the one electron states: | |||
:{| cellpadding="5" cellspacing="0" border="1" | |||
|{{TAG|LORBIT}}=10|| {{TAG|DOSCAR}} and l decomposed {{TAG|PROCAR}} file | |||
|- | |||
|{{TAG|LORBIT}}=11|| {{TAG|DOSCAR}} and lm decomposed {{TAG|PROCAR}} file | |||
|} | |||
*We'll use {{TAG|LORBIT}}=11 and see if we can distinguish <math>p_{x}</math> and <math>p_{z}</math> states. | |||
=== {{TAG|PROCAR}} === | |||
band 3 # energy -11.46540832 # occ. 2.00000000 | |||
ion s py pz px dxy dyz dz2 dxz dx2 tot | |||
1 0.000 0.510 0.000 0.036 0.000 0.000 0.000 0.000 0.000 0.546 | |||
2 0.000 0.146 0.000 0.010 0.000 0.000 0.000 0.000 0.000 0.157 | |||
tot 0.000 0.656 0.000 0.047 0.000 0.000 0.000 0.000 0.000 0.703 | |||
band 4 # energy -11.46540832 # occ. 2.00000000 | |||
ion s py pz px dxy dyz dz2 dxz dx2 tot | |||
1 0.000 0.036 0.000 0.510 0.000 0.000 0.000 0.000 0.000 0.546 | |||
2 0.000 0.010 0.000 0.146 0.000 0.000 0.000 0.000 0.000 0.157 | |||
tot 0.000 0.047 0.000 0.656 0.000 0.000 0.000 0.000 0.000 0.703 | |||
band 5 # energy -8.76483386 # occ. 2.00000000 | |||
ion s py pz px dxy dyz dz2 dxz dx2 tot | |||
1 0.001 0.000 0.135 0.000 0.000 0.000 0.000 0.000 0.000 0.136 | |||
2 0.172 0.000 0.261 0.000 0.000 0.000 0.000 0.000 0.000 0.433 | |||
tot 0.173 0.000 0.396 0.000 0.000 0.000 0.000 0.000 0.000 0.569 | |||
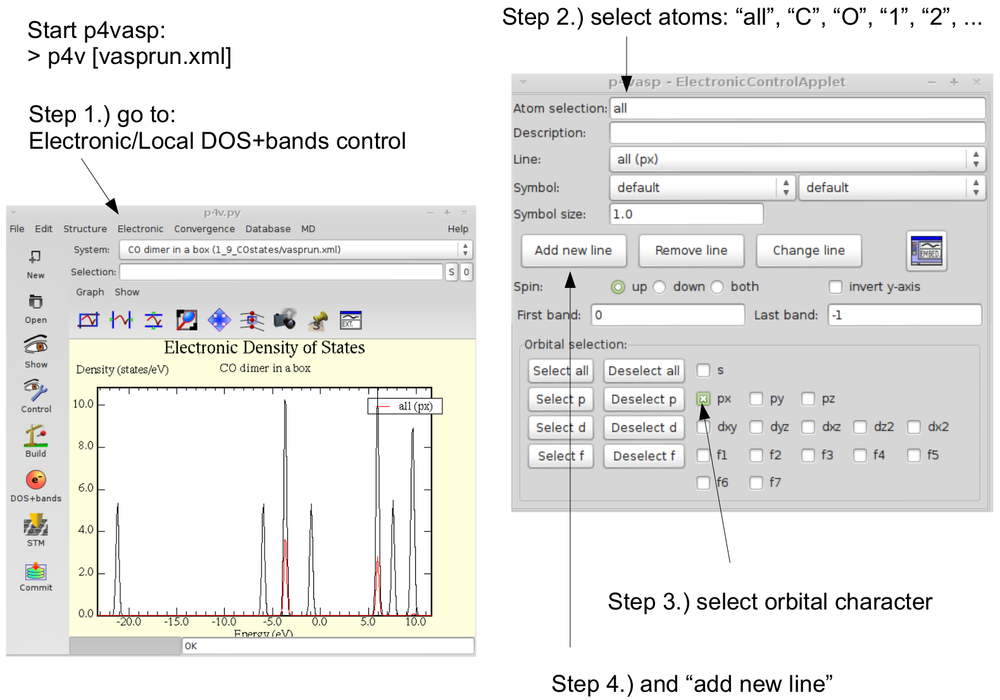
*To plot the DOS start p4vasp: | |||
[[File:Fig CO 1.png|1000px]] | |||
== Download == | == Download == | ||
[ | [[Media:COstates.tgz| COstates.tgz]] | ||
{{Template:At_and_mol}} | |||
[[Category:Examples]] | [[Category:Examples]] | ||
Latest revision as of 13:19, 14 November 2019
Overview > O atom > O atom spinpolarized > O atom spinpolarized low symmetry > O dimer > CO > CO vibration > CO partial DOS > H2O >
H2O vibration > H2O molecular dynamics > Further things to try > List of tutorials
Task
Calculation of the DOS of a CO molecule (using p4vasp)
Input
POSCAR
CO molecule in a box 1.0 ! universal scaling parameters 8.0 0.0 0.0 ! lattice vector a(1) 0.0 8.0 0.0 ! lattice vector a(2) 0.0 0.0 8.0 ! lattice vector a(3) 1 1 ! number of atoms for each species sel ! selective degrees of freedom are changed cart ! positions in cartesian coordinates 0 0 0 F F T ! first atom 0 0 1.143 F F T ! second atom
INCAR
SYSTEM = CO molecule in a box ISMEAR = 0 ! Gaussian smearing LORBIT = 11
KPOINTS
Gamma-point only 0 Monkhorst Pack 1 1 1 0 0 0
Calculation
- The PROCAR file gives valuable information of the character of the one electron states:
- We'll use LORBIT=11 and see if we can distinguish and states.
PROCAR
band 3 # energy -11.46540832 # occ. 2.00000000 ion s py pz px dxy dyz dz2 dxz dx2 tot 1 0.000 0.510 0.000 0.036 0.000 0.000 0.000 0.000 0.000 0.546 2 0.000 0.146 0.000 0.010 0.000 0.000 0.000 0.000 0.000 0.157 tot 0.000 0.656 0.000 0.047 0.000 0.000 0.000 0.000 0.000 0.703 band 4 # energy -11.46540832 # occ. 2.00000000 ion s py pz px dxy dyz dz2 dxz dx2 tot 1 0.000 0.036 0.000 0.510 0.000 0.000 0.000 0.000 0.000 0.546 2 0.000 0.010 0.000 0.146 0.000 0.000 0.000 0.000 0.000 0.157 tot 0.000 0.047 0.000 0.656 0.000 0.000 0.000 0.000 0.000 0.703
band 5 # energy -8.76483386 # occ. 2.00000000 ion s py pz px dxy dyz dz2 dxz dx2 tot 1 0.001 0.000 0.135 0.000 0.000 0.000 0.000 0.000 0.000 0.136 2 0.172 0.000 0.261 0.000 0.000 0.000 0.000 0.000 0.000 0.433 tot 0.173 0.000 0.396 0.000 0.000 0.000 0.000 0.000 0.000 0.569
- To plot the DOS start p4vasp: